Project No. 2457
STANDARD PROJECT
Primary Supervisor
Dr Samuel Robson – University of Portsmouth
Co-Supervisor(s)
Dr Benjamin Towler – University of Sussex
Dr Garry Scarlett – University of Portsmouth
Summary
The i-motif is a cytosine rich, non-standard nucleotide structure, very different from the typical Watson-Crick double-helical structure of DNA.
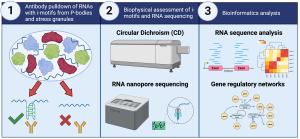
Although its existence has been established since the 1990s, it was previously believed that the i-motif structure was not biologically relevant due to its preference for low pH. However, recent evidence has directly visualised i-motif structures in the genomes of living cells. This, along with other recent evidence, shows that i-motifs not only exist in genomes but also play a biological role. Through the use of an i-motif specific antibody (i-Mab), our team have generated one of the first whole-genome maps of biologically stable i-motifs in DNA, using a combined approach of CUT&RUN and next generation sequencing (NGS). However, whilst formation in DNA represents a likely regulatory mechanism for gene expression, their role in control of expression at the RNA transcript level remains to be seen.
In this project, we will further explore the role of i-motifs in regulation in the context of their formation in RNA, in particular with respect to their prevalence within processing bodies (P-bodies) and stress granules. The project will pursue similar approaches to those that we have employed for DNA: first using the i-Mab antibody to select for RNAs containing the i-motif structure; biophysical and biochemical validation of the selected targets; and finally bioinformatic comparison of i-motif containing sequences to generate new insights into the role of this structure at the transcript level. This project represents an exciting opportunity for the candidate to work in a number of cutting-edge fields, and develop skills in RNA nanopore sequencing, bioinformatics, biophysics, and RNA biology.
The ideal candidate will have experience in both wet lab biology and computational approaches, show an aptitude for developing new skills, and demonstrate an acute sense of curiosity about the functionality of life and how cells work.