Project No. 2469
STANDARD PROJECT
Primary Supervisor
Assoc Prof Eugen Stulz – University of Southampton
Co-Supervisor(s)
Dr David Rusling – University of Portsmouth
Prof Sean Lim – University of Southampton
Summary
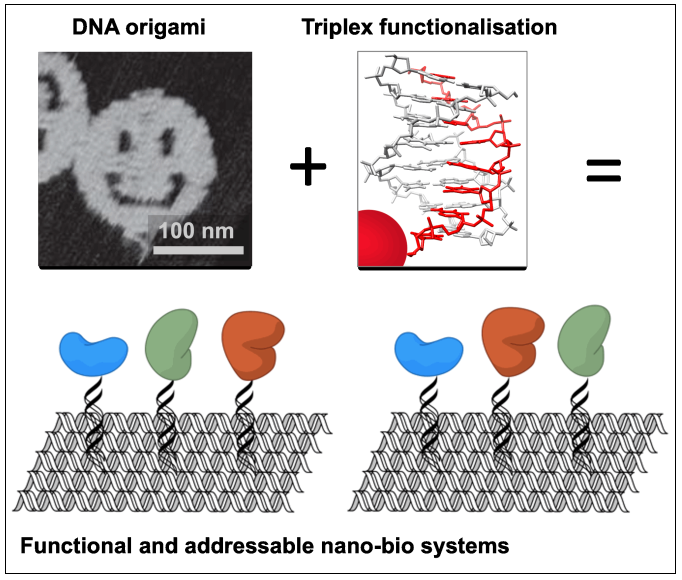
DNA origami has proven to be a versatile approach for the directed self-assembly of custom objects bearing designed features on the nanometre scale
Functionalising these structures with non-nucleic acid components such as proteins/enzymes offers applications in the study of biological cascades, the structure determination of periodically positioned molecules, or sensing. To date, the introduction of biomolecules has relied on their direct attachment to the underlying DNA which influences folding of the structures, and is not compatible with most biological molecules which are denatured during annealing.
To address this problem, we have developed the triplex approach to DNA recognition to introduce components into DNA nanostructures after origami assembly, by exploiting the sequence addressability of the underlying double-helix of the origami. Triplex-forming oligonucleotides (TFOs) are short nucleic acid strands that bind sequence specifically within the duplex major groove at room temperature, generating a triple-helical structure. TFOs can be programmed to target unique positions within a DNA nanostructure and allow the introduction of components by their covalent attachment to the TFO ends. By using synthetic nucleotides, this works now well at neutral pH. So far much of our work has focused on simple tile-based structures; this project will extend this approach to more complex DNA origami (2D and 3D objects, arrays…) and various types of biomolecules that can be introduced.
We will focus on non-canonical DNA structures, proteins/enzymes, and mono-clonal antibodies, to create platforms for analysing their biological activity in an oriented manner. Specific attachment to biomolecules via click-chemistry is available. A major part of the project will focus on the spectroscopic characterisation and activity testing (enzyme assays, binding to aptamers and antibodies) of the constructs – how does specific orientation affect activity?
The ideal candidate will have a good background in organic chemistry and show enthusiasm to work with biological systems. This project is where nano meets biology meets chemistry; the training will be interdisciplinary crossing many boundaries, and will give the student a wealth of highly sought-after skills.