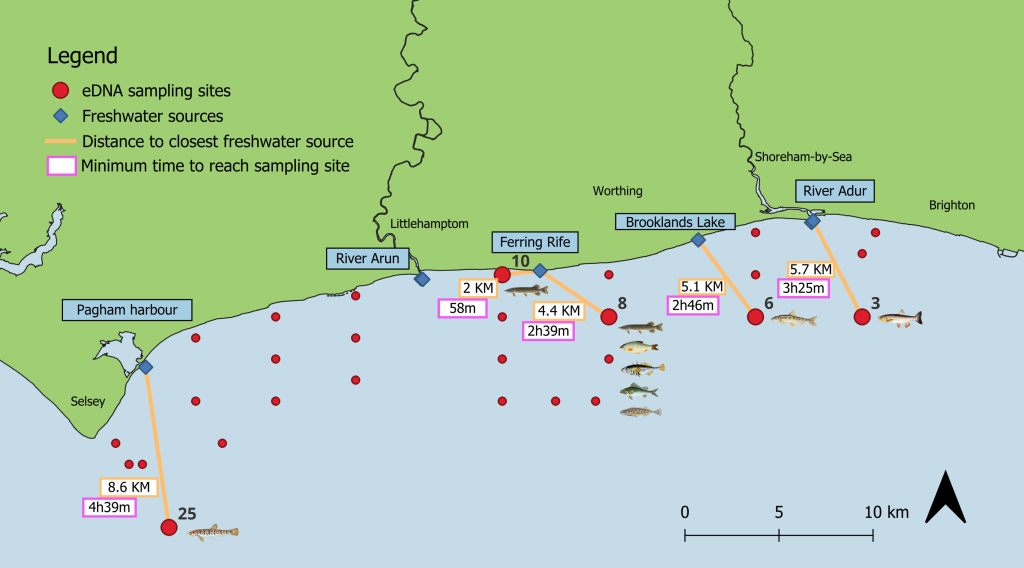
Environmental DNA (eDNA) has gained popularity in the last decade as an effective biomonitoring tool in aquatic environments. eDNA consists of genetic material shed by organisms into the environment through scales, skin cells, faeces and other excrements. It is a non-invasive, relatively affordable, and time-effective technique that eliminates the need for taxonomic expertise to identify species. One key advantage of eDNA metabarcoding is its ability to detect a wide range of taxa simultaneously, providing a snapshot of the species present within an ecosystem at a given time, and has been shown to be more sensitive than traditional biomonitoring methods. More recently, the use of eDNA to indirectly estimate marine species abundance has been gaining momentum. However, several factors are known to influence the ability of eDNA to reflect relative abundance.
In a recent publication, final year SoCoBio student Alice Clark conducted a study to investigate the feasibility of using eDNA metabarcoding to estimate relative abundance of marine species in a dynamic tidal environment, using Sussex Bay, UK, as a case study.
Read the publication in full: https://doi.org/10.1002/edn3.70131